1. Pharmacological Profiling (A-C. Camproux & O. Taboureau, resp, A. Badel, D. Flatters, C. Geneix, M. Petitjean, L. Regad):
Keywords: polypharmacology, protein-small molecule interactions, target and binding site structural analysis, biostatistics, bioinformatics, chemoinformatics, data integration, systems pharmacology and toxicology, network biology.
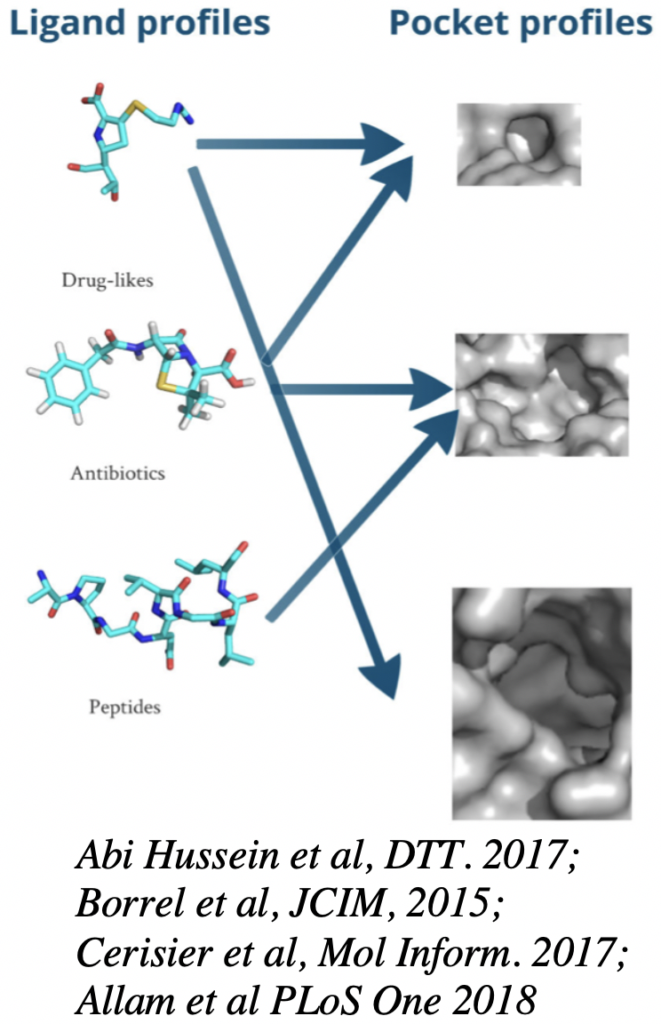
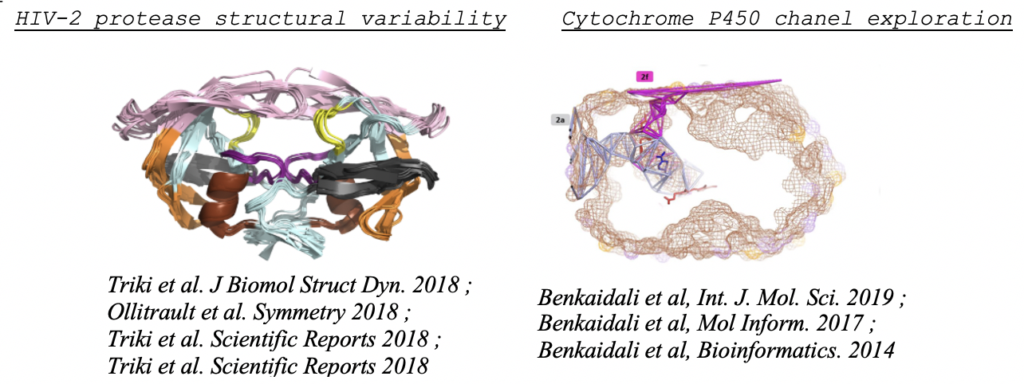
– Structural analyses of protein targets and prediction of target-ligand interactions. The objective is to better understand and predict interactions between a drug and a target. We perform structural analyses of the target structure by considering its flexibility in order to understand the deformation induced by drug interactions. We characterize drug and target binding-site properties and their corresponding profiles in the aim to predict their interactions, using chemoinformatics, molecular dynamics simulations and structural bioinformatics methods. We develop protocols to explore drug-target interactions by i) studying the impact of mutations on the target structure and its interactions with drugs and ii) developing biostatistical methods for the prediction of drug-target interactions. We also explore off-target prediction and promiscuity effects. This research is applied on the protease enzyme of HIV-2, the proteins NS1/NS2 of influenza and to enzymes and toxicology targets, in collaboration with experimentalists.
– Molecular and structural modelling and virtual screening to discover new hits candidates, to determine mechanism of binding and to evaluate the pharmacological profile of a small molecule. We work for example on metabolite enzymes (CBS, Dirk1A) that play a role in Down syndrome.
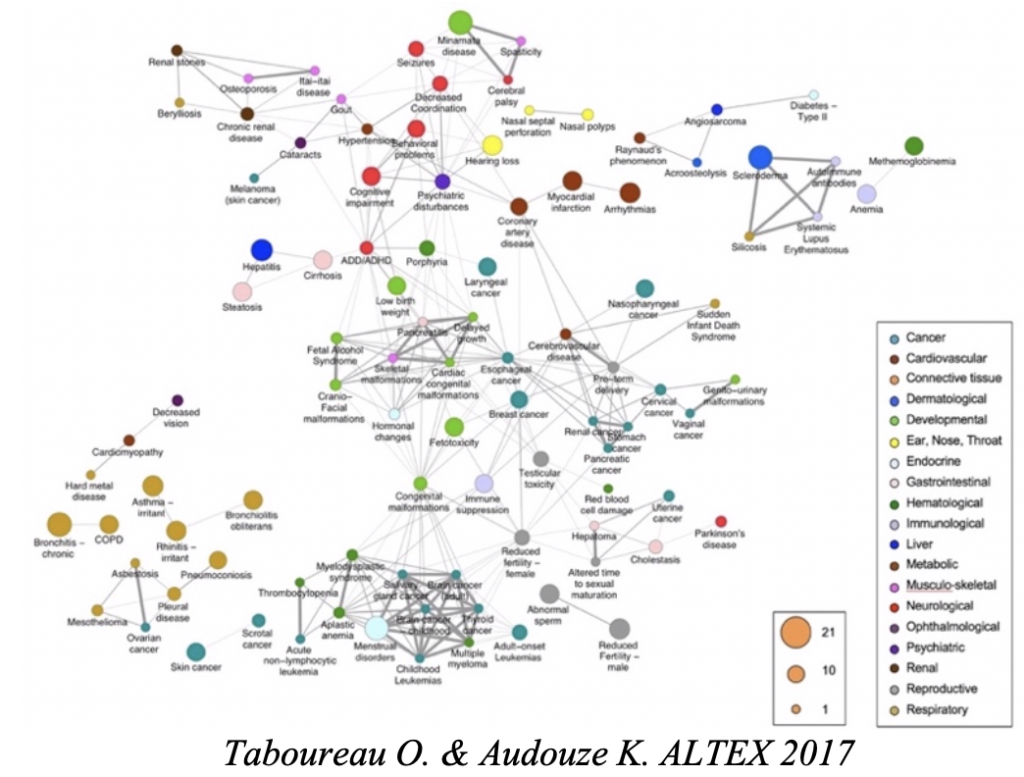
– Analysis of the ligand-phenotype relationship. The objective is to analyze the impact of small molecules and chemical exposure to human at different levels of complexity (from molecular, cellular and tissue) that could lead to adverse effect and diseases. We develop integrative systems pharmacology and toxicology approaches using network biology, biostatistics and transcriptomics data analysis that could be relevant for the identification of adverse outcome pathways (AOPs) and mechanisms involved in some pathologies such as drug-induced liver injury (DILI), steatosis, diabetes or reproduction disorders.
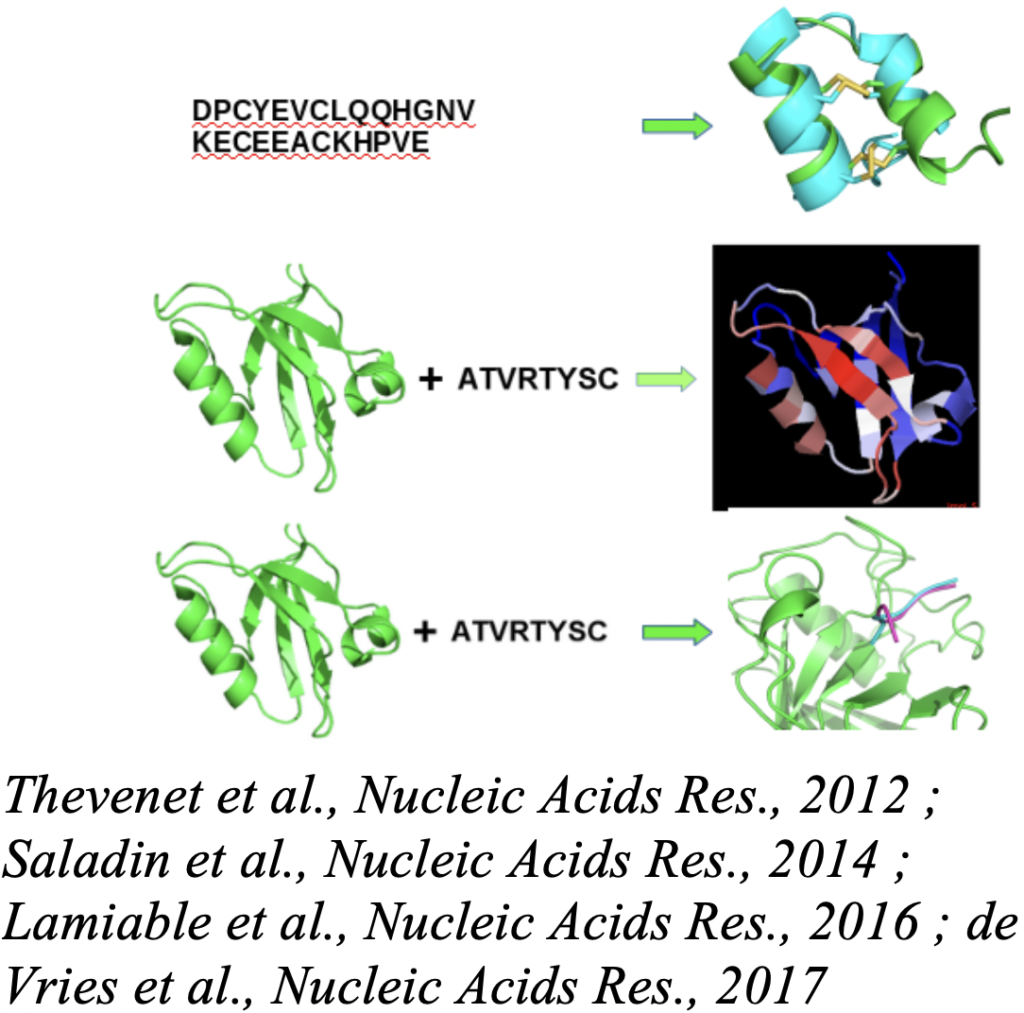
2. Peptide-Protein Interactions and Design of Peptide Probes or Candidate Peptides (P. Tufféry, resp., G. Moroy, S. Murail, J. Rey, S. De Vries):
Keywords : Protein peptide interactions, therapeutic peptide design, methodology, structure based peptide design, interfering peptides.
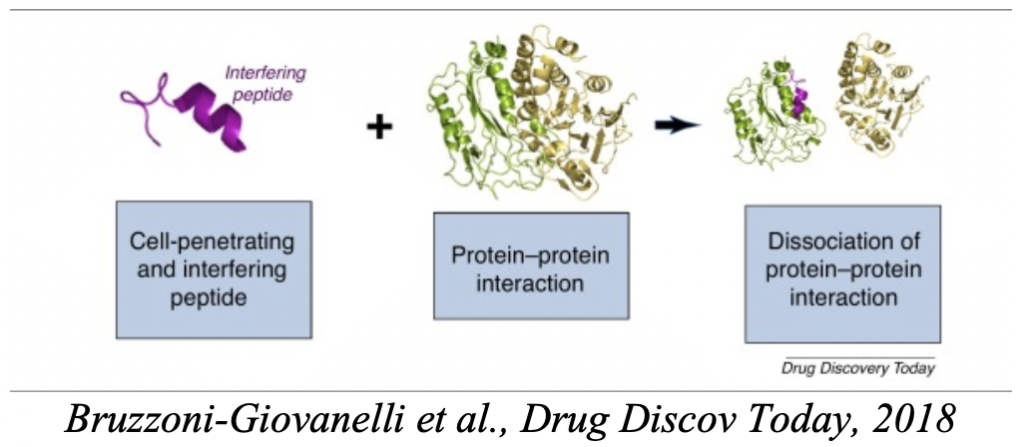
Our main – but not exclusive – field of interest is the design of interfering peptides targeting specific protein-protein interactions, in the context of metabolism regulation, or cancer deregulation. This activity is supported by methodological developments on the following directions :
– The design of peptide probes or candidate peptides, which implies de development of specific in silico protocols. We study peptide protein interactions driven by the knowledge of the protein structure and have developped tools such as PEP-FOLD, PEP-SiteFinder or pepATTRACT, able to predict peptide structure in isolation of in interaction with its receptor, and to identify candidate binding sites on the receptor structure. Current efforts are on peptide-protein docking (hot spots, conformational sampling, scoring), and on the development of approaches to optimize peptides (cyclization, sequence variants). We are also interested in coupling in silico with the PEPscan in vitro approach to the identification of peptides interfering with target protein-protein interactions.
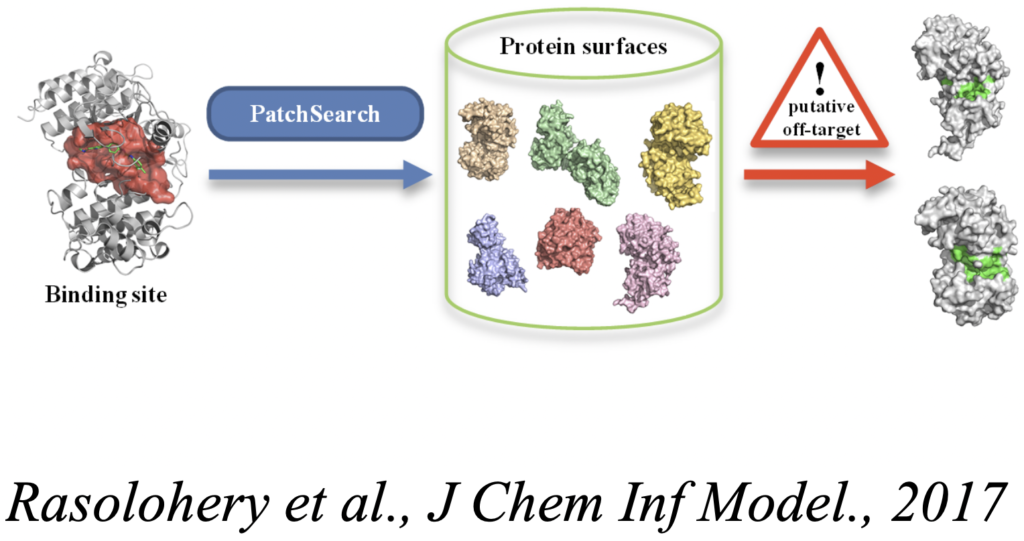 – Knowledge inference using structural data mining. The large scale analysis of the increasing collection of protein structures solved experimentally has high potential for the the study of interaction selectivity and the development of peptide specific of target protein protein interactions. We are interested in quantifying the similarity of both protein fragment conformations (see BCSearch, DaReUs-Loop) and surface patches (PatchSearch), and their application to the design of peptides specific of targets.
– Knowledge inference using structural data mining. The large scale analysis of the increasing collection of protein structures solved experimentally has high potential for the the study of interaction selectivity and the development of peptide specific of target protein protein interactions. We are interested in quantifying the similarity of both protein fragment conformations (see BCSearch, DaReUs-Loop) and surface patches (PatchSearch), and their application to the design of peptides specific of targets.
3. RPBS Platform (“Ressource Parisienne en Bioinformatique Structurale”):
Keywords : Structural bioinformatics, chemoinformatics.
We host and administrate the RPBS platform (bioserv.rpbs.univ-paris-diderot.fr) a national platform focusing on (i) structural bioinformatics : analysis and modeling of protein structure and protein-protein interactions, and (ii) the identification of therapeutic compounds (docking/screening of small compounds, docking/optimization of peptides). RPBS is an IbiSA platform member of the French Institute for Bioinformatics (IFB – www.france-bioinformatique.fr) and of ChemBioFrance (www.chembiofrance.fr).